Importing a VCF into Bioconductor
Overview
Teaching: 45 min
Exercises: 15 minQuestions
How can I import and filter a VCF with Bioconductor?
Objectives
Read genotypes and SNP metadata from a VCF into R.
Compress and index a VCF for quick access.
Use custom parameters to filter variants into a smaller file.
Choose which fields, samples, and genomic ranges to import into R.
library(GenomicRanges)
library(Rsamtools)
library(VariantAnnotation)
library(magrittr)
Making the VCF file easy to access
We have a small VCF for this workshop, but they can be many gigabytes in size. So, instead of reading straight from a plain text file, we will compress and index the file. This only needs to be done to the file once, even if you will read data from the file many times in later R sessions.
To save time, the file you downloaded was zipped already. If it wasn’t, you would zip it this way:
bg <- bgzip("data/hmp321_agpv4_chr1_subset.vcf")
BGZIP format
The BGZIP format is an extension of the GZIP format, so any command that would work on
.gzfiles (likezless) works on.bgzfiles. The BGZIP format was designed specifically for bioinformatics files to make them easy to index. See http://www.htslib.org/doc/bgzip.html for more information.
Instead we’ll input the zipped file name directly.
bg <- "data/hmp321_agpv4_chr1_subset.vcf.bgz"
Now we’ll build an index that will make it easy for R to read just particular regions of the genome.
indexTabix(bg, format = "vcf")
[1] "data/hmp321_agpv4_chr1_subset.vcf.bgz.tbi"
Now we can make an object that points to the file for quick access, kind of like
the FaFile object we made in Episode 2.
myvcf <- VcfFile(bg)
Scanning the header
Even a VCF that is small when compressed can take up a lot of memory when imported into R. So, we’ll want to think carefully about how we import it. One helpful thing you can do is just read the header, to learn more about what data the VCF contains.
hdr <- scanVcfHeader(myvcf)
hdr
class: VCFHeader
samples(200): 2005-4 207 ... german_F03802 german_EZ5
meta(4): HapMapVersion fileDate fileformat contig
fixed(2): FILTER ALT
info(18): DP NZ ... ImpMinorAccuracy DUP
geno(3): GT AD GL
Let’s go through the different components that were listed.
With samples, we can get a character vector of all sample names in the file.
This allows us to confirm that the file contains the samples that we expect it
to. Later we can also decide to only import genotypes from certain samples.
all_sam <- samples(hdr)
all_sam
[1] "2005-4" "207" "32"
[4] "5023" "680" "68139"
[7] "697" "75-14gao" "764"
[10] "78004" "78551S" "792"
[13] "83IBI3" "8982" "9058"
[16] "98F1" "B4" "B7"
[19] "B73" "B76" "B8"
[22] "B97" "BKN009" "BKN011"
[25] "BKN017" "BKN018" "BKN027"
[28] "BKN029" "C521" "CAUMo17"
[31] "chang69" "chang7daxian1" "CML103"
[34] "CML103-run248" "CML411" "CN104"
[37] "Co109" "CT109" "D1139"
[40] "D20" "D23" "D801"
[43] "D857" "D892" "dai6"
[46] "DM07" "dupl-478" "E588"
[49] "E601" "F7584" "F939"
[52] "FR14" "fu8538" "H114"
[55] "H84" "HD568" "hua160"
[58] "huangchanga" "huotanghuang17" "Il14H"
[61] "ji63" "K22" "Ki11"
[64] "Ky21" "LD61" "LH1"
[67] "LH128" "LH190" "LH202"
[70] "LH51" "LH60" "lian87"
[73] "liao2204" "LP1" "luyuan133"
[76] "Lx9801" "M3736" "MBUB"
[79] "Ms71" "mu6" "N138"
[82] "N192" "N42" "NC268"
[85] "NC358" "ning24" "ning45"
[88] "NS501" "Oh40B" "Pa91"
[91] "PHG50" "PHG83" "PHJ31"
[94] "PHJ75" "PHK05" "PHM10"
[97] "PHN11" "PHNV9" "PHP02"
[100] "PHR58" "PHT77" "PHW17"
[103] "PHW52" "PHWG5" "qiong51"
[106] "R1656" "RS710" "SC24-1"
[109] "SG17" "shangyin110-1" "shen142"
[112] "SS99" "SZ3" "tai184"
[115] "TIL01-JD" "TIL03" "TIL09"
[118] "Timpunia-1" "VL056883" "VL062784"
[121] "W117" "W238" "W344"
[124] "W499" "W668" "W968"
[127] "W9706" "wenhuang31413" "WIL900"
[130] "wu312" "XF117" "y9961"
[133] "yan38" "Yd6" "ye107"
[136] "ye8112" "yue39-4" "yue89A12-1"
[139] "zhangjin6" "MAIdgiRAPDIAAPEI-12" "MAIdgiRAVDIAAPEI-4"
[142] "MAIdgiRCKDIAAPEI-9" "ZEAhwcRAXDIAAPE" "ZEAxppRALDIAAPEI-9"
[145] "ZEAxppRAUDIAAPEI-1" "ZEAxppRBFDIAAPEI-3" "ZEAxppRBMDIAAPEI-6"
[148] "ZEAxppRCODIAAPEI-9" "ZEAxppRDLDIAAPEI-2" "ZEAxujRBADIAAPE"
[151] "282set_A556" "282set_A619" "282set_A634"
[154] "282set_A654" "282set_A659" "282set_A661"
[157] "282set_A679" "282set_B103" "282set_CH701-30"
[160] "282set_CI187-2" "282set_CI31A" "282set_CI64"
[163] "282set_CM7" "282set_CML14" "282set_CML154Q"
[166] "282set_CML254" "282set_CML287" "282set_GT112"
[169] "282set_H99" "282set_I29" "282set_IDS28"
[172] "282set_Ki21" "282set_KY228" "282set_MS153"
[175] "282set_Mt42" "282set_NC222" "282set_NC264"
[178] "282set_NC338" "282set_NC346" "282set_NC360"
[181] "282set_NC366" "282set_OH7B" "282set_Os420"
[184] "282set_Pa875" "282set_Sg1533" "282set_T232"
[187] "282set_T234" "282set_Tx601" "282set_Tzi25"
[190] "282set_Tzi8" "282set_VA102" "282set_Va14"
[193] "282set_Va26" "282set_W117Ht" "282set_Wf9"
[196] "german_Mo17" "german_Lo11" "german_FF0721H-7"
[199] "german_F03802" "german_EZ5"
We can see the file date and other general information from the top of the file.
meta(hdr)
DataFrameList of length 4
names(4): HapMapVersion fileDate fileformat contig
for(i in names(meta(hdr))){
meta(hdr)[[i]] %>% show()
}
DataFrame with 1 row and 1 column
Value
<character>
HapMapVersion "3.2.1"
DataFrame with 1 row and 1 column
Value
<character>
fileDate 20201110
DataFrame with 1 row and 1 column
Value
<character>
fileformat VCFv4.1
DataFrame with 1 row and 1 column
assembly
<character>
1 "1"
For contig we only see "1" because this file only contains variants on
chromosome 1.
We can print out descriptions of all the INFO fields.
info(hdr)
DataFrame with 18 rows and 3 columns
Number Type Description
<character> <character> <character>
DP 1 Integer Total Depth
NZ 1 Integer Number of taxa with ..
AD . Integer Total allelelic dept..
AC . Integer Numbers of ALT allel..
AQ . Integer Average phred base q..
... ... ... ...
NI5 0 Flag Site with 5bp of a p..
INHMP311 0 Flag Site peresent in Hap..
ImpHomoAccuracy 1 Float Fraction of homozygo..
ImpMinorAccuracy 1 Float Fraction of minor al..
DUP 0 Flag Site with heterozygo..
info_desc <- info(hdr)$Description
names(info_desc) <- rownames(info(hdr))
info_desc
DP
"Total Depth"
NZ
"Number of taxa with called genotypes"
AD
"Total allelelic depths in order listed starting with REF"
AC
"Numbers of ALT alleles in order listed"
AQ
"Average phred base quality for alleles in order listed starting with REF"
GN
"Number of taxa with genotypes AA,AB,BB or AA,AB,AC,BB,BC,CC if 2 alt alleles"
HT
"Number of heterozygotes"
EF
"EF=het_frequency/(presence_frequency * minor_allele_frequency), if 2 alt alleles,EF for AB,AC,BC pairsis given; from 916 taxa of HapMap 3.1.1"
PV
"p-value from segregation test between AB or AB, AC, BC if 2 alt alleles, from 916 taxa of HapMap 3.1.1"
MAF
"Minor allele frequency"
MAF0
"Minor allele frequency from unimputed HapMap 3.2.1 on 1210 taxa"
IBD1
"only one allele present in IBD contrasts; based on 916 taxa of HapMap3.1.1"
LLD
"Site in local LD with GBS map (from 916 taxa of HapMap 3.1.1)"
NI5
"Site with 5bp of a putative indel from 916 taxa of HapMap 3.1.1"
INHMP311
"Site peresent in HapMap3.1.1"
ImpHomoAccuracy
"Fraction of homozygotes imputed back into homozygotes"
ImpMinorAccuracy
"Fraction of minor allele homozygotes imputed back into minor allelel homozygotes"
DUP
"Site with heterozygotes frequency > 3%"
Descriptions of the FORMAT fields
Print out descriptions of the
FORMATfields. Look athdragain to find a function to do this. You might also look back at Episode 1 to remind yourself of what theFORMATfields represent.Solution
geno(hdr)DataFrame with 3 rows and 3 columns Number Type Description <character> <character> <character> GT 1 String Genotype AD . Integer Allelic depths for t.. GL . Integer Genotype likelihoods..geno(hdr)$Description[1] "Genotype" [2] "Allelic depths for the reference and alternate alleles in the order listed" [3] "Genotype likelihoods for 0/0, 0/1, 1/1, or 0/0, 0/1, 0/2, 1/1, 1/2, 2/2 if 2 alt alleles"
Importing the first chunk of the file
How are the actual data formatted? We don’t want to read the whole file, but here is how we can read just the first 100 entries.
myvcf2 <- VcfFile(bg, yieldSize = 100)
test <- readVcf(myvcf2)
test
class: CollapsedVCF
dim: 100 200
rowRanges(vcf):
GRanges with 5 metadata columns: paramRangeID, REF, ALT, QUAL, FILTER
info(vcf):
DataFrame with 18 columns: DP, NZ, AD, AC, AQ, GN, HT, EF, PV, MAF, MAF0, ...
info(header(vcf)):
Number Type Description
DP 1 Integer Total Depth
NZ 1 Integer Number of taxa with called genotypes
AD . Integer Total allelelic depths in order listed st...
AC . Integer Numbers of ALT alleles in order listed
AQ . Integer Average phred base quality for alleles in...
GN . Integer Number of taxa with genotypes AA,AB,BB or...
HT 1 Integer Number of heterozygotes
EF . Float EF=het_frequency/(presence_frequency * mi...
PV . Float p-value from segregation test between AB ...
MAF 1 Float Minor allele frequency
MAF0 1 Float Minor allele frequency from unimputed Hap...
IBD1 0 Flag only one allele present in IBD contrasts;...
LLD 0 Flag Site in local LD with GBS map (from 916 t...
NI5 0 Flag Site with 5bp of a putative indel from 91...
INHMP311 0 Flag Site peresent in HapMap3.1.1
ImpHomoAccuracy 1 Float Fraction of homozygotes imputed back into...
ImpMinorAccuracy 1 Float Fraction of minor allele homozygotes impu...
DUP 0 Flag Site with heterozygotes frequency > 3%
geno(vcf):
List of length 3: GT, AD, GL
geno(header(vcf)):
Number Type Description
GT 1 String Genotype
AD . Integer Allelic depths for the reference and alternate alleles ...
GL . Integer Genotype likelihoods for 0/0, 0/1, 1/1, or 0/0, 0/1, 0...
We could get the same header from above using the header function.
We can also now get information about the first 100 variants. The rowRanges
function gets us a GRanges object indicating the name and position of each
SNP, along with the reference and alternative allele.
rowRanges(test)
GRanges object with 100 ranges and 5 metadata columns:
seqnames ranges strand | paramRangeID REF
<Rle> <IRanges> <Rle> | <factor> <DNAStringSet>
1-20689192 1 21000162 * | NA G
1-20689220 1 21000190 * | NA G
1-20689236 1 21000206 * | NA C
1-20689304 1 21000274 * | NA C
1-20689307 1 21000277 * | NA A
... ... ... ... . ... ...
1-20690571 1 21001541 * | NA C
1-20690577 1 21001547 * | NA C
1-20690617 1 21001587 * | NA C
1-20690618 1 21001588 * | NA G
1-20690639 1 21001609 * | NA A
ALT QUAL FILTER
<CharacterList> <numeric> <character>
1-20689192 T NA PASS
1-20689220 A NA PASS
1-20689236 A NA PASS
1-20689304 A NA PASS
1-20689307 G NA PASS
... ... ... ...
1-20690571 T NA PASS
1-20690577 A NA PASS
1-20690617 T NA PASS
1-20690618 A NA PASS
1-20690639 T NA PASS
-------
seqinfo: 1 sequence from "1" genome; no seqlengths
The info function gets us a data frame with all of the INFO variables.
info(test)
DataFrame with 100 rows and 18 columns
DP NZ AD AC AQ
<integer> <integer> <IntegerList> <IntegerList> <IntegerList>
1-20689192 853 1203 844,9 14 34,34
1-20689220 857 1203 847,10 15 33,32
1-20689236 866 1190 674,192 193 33,30
1-20689304 961 1181 878,83 153 35,32
1-20689307 961 1210 957,4 4 33,30
... ... ... ... ... ...
1-20690571 3314 1210 3281,33 6 34,35
1-20690577 3199 1182 599,2600 1829 34,33
1-20690617 3289 1210 3277,12 4 35,37
1-20690618 3271 1210 3251,20 10 35,35
1-20690639 3256 1210 3252,4 2 34,32
GN HT EF PV MAF
<IntegerList> <integer> <NumericList> <NumericList> <numeric>
1-20689192 1195,2,6 2 1 0.001 0.006
1-20689220 1194,3,6 3 1.23 0.002 0.006
1-20689236 1085,17,88 17 0.71 0 0.081
1-20689304 1098,13,70 13 1.59 0 0.065
1-20689307 1207,2,1 2 2.24 0.004 0.002
... ... ... ... ... ...
1-20690571 1206,2,2 2 1.45 0 0.002
1-20690577 266,3,913 3 0.06 0 0.226
1-20690617 1208,0,2 0 0 0 0.002
1-20690618 1204,2,4 2 0.82 0 0.004
1-20690639 1209,0,1 0 0 0.008 0.001
MAF0 IBD1 LLD NI5 INHMP311 ImpHomoAccuracy
<numeric> <logical> <logical> <logical> <logical> <numeric>
1-20689192 0.020 TRUE FALSE FALSE FALSE 0.983942
1-20689220 0.019 FALSE FALSE FALSE FALSE 0.988387
1-20689236 0.167 FALSE TRUE FALSE TRUE 0.970551
1-20689304 0.068 FALSE TRUE FALSE TRUE 0.981187
1-20689307 0.004 TRUE FALSE FALSE FALSE 0.995828
... ... ... ... ... ... ...
1-20690571 0.005 TRUE FALSE FALSE FALSE 0.999100
1-20690577 0.205 FALSE TRUE FALSE TRUE 0.995400
1-20690617 0.003 TRUE FALSE FALSE FALSE 0.995816
1-20690618 0.008 FALSE FALSE FALSE FALSE 0.995766
1-20690639 0.002 TRUE FALSE FALSE TRUE 0.997494
ImpMinorAccuracy DUP
<numeric> <logical>
1-20689192 0.000000 FALSE
1-20689220 0.307692 FALSE
1-20689236 0.859813 TRUE
1-20689304 0.962963 TRUE
1-20689307 0.000000 FALSE
... ... ...
1-20690571 0.800000 FALSE
1-20690577 0.977578 FALSE
1-20690617 0.000000 FALSE
1-20690618 0.444444 FALSE
1-20690639 0.000000 FALSE
The geno function gets us data associated with each genotype. We use the $
accessor to get each field.
geno(test)$GT[1:10,1:5]
2005-4 207 32 5023 680
1-20689192 "0/0" "0/0" "0/0" "0/0" "0/0"
1-20689220 "0/0" "0/0" "0/0" "0/0" "0/0"
1-20689236 "0/0" "0/0" "0/0" "0/0" "0/0"
1-20689304 "0/0" "0/0" "0/0" "0/0" "0/0"
1-20689307 "0/0" "0/0" "0/0" "0/0" "0/0"
1-20689321 "0/0" "0/0" "0/0" "0/0" "0/0"
1-20689326 "0/0" "0/0" "0/0" "1/1" "./."
1-20689334 "0/0" "0/0" "0/0" "0/0" "0/0"
1-20689549 "0/0" "0/0" "0/0" "0/0" "0/0"
1-20689561 "1/1" "1/1" "./." "0/0" "1/1"
Reading the file in chunks
With the
yieldSizeargument, we could actually callreadVcfrepeatedly to go through the whole file one chunk at a time. In that case we would callopen(myvcf2)before callingreadVcf, then callclose(myvcf2)once we were done reading the file.
Filtering the file
Before we import, we can filter the file to eliminate variants that we know we aren’t interested in. This is optional of course, but will save us some memory and computational time when we import the data later. It will also give us a VCF that we could import into other software.
Let’s filter on a few of the INFO fields. According to
documentation of this dataset, the LLD
flag indicates good quality markers. The DUP flag indicates a high proportion
of heterozygotes (for an inbred crop like maize) and so we should probably throw
away markers with that flag. Finally, let’s set a minimum minor allele frequency
(MAF) at 0.01. We’ll look at the formatting of these fields.
info(hdr)[c("LLD", "DUP", "MAF"),]
DataFrame with 3 rows and 3 columns
Number Type Description
<character> <character> <character>
LLD 0 Flag Site in local LD wit..
DUP 0 Flag Site with heterozygo..
MAF 1 Float Minor allele frequency
LLD and DUP are “flags”, which means that the abbreviation will either be
present or absent in the INFO column. MAF is a “float” (which just means
a number that is not necessarily an integer) with one value per marker.
For each rule, we need to create a function that will return TRUE if we want
to keep the variant and FALSE if we want to discard it. For our MAF filter,
we’ll make a function that can work on an imported VCF.
Creating functions
See the software carpentry tutorial on how to create functions in R.
maf01 <- function(vcf){
info(vcf)$MAF >= 0.01
}
We can test it out on the chunk of the VCF we imported above.
maf01(test) %>% head()
[1] FALSE FALSE TRUE TRUE FALSE TRUE
When we can, it’s more efficient for filtering to test the plain text of the
file than to import it into R. Because LLD and DUP are “flags”, those
abbreviations will either be in the INFO field or not, which translates
to TRUE and FALSE in our info(test) data frame. We can use the grepl
function to search for those strings in the unprocessed file. Let’s use
readLines to import some of the file for testing.
testlines <- readLines(bg, n = 150)
testlines <- testlines[!startsWith(testlines, "#")] # eliminate headers
Now we’ll test using grepl. For space I am just looking at two lines, but
in real life I would probably look at more.
testLLD <- grepl("LLD", testlines)
head(testlines[testLLD], n = 2) # lines that should have LLD
[1] "1\t21000206\t1-20689236\tC\tA\t.\tPASS\tDP=866;NZ=1190;AD=674,192;AC=193;AQ=33,30;GN=1085,17,88;HT=17;EF=0.71;PV=0;MAF=0.081;MAF0=0.167;LLD;INHMP311;ImpHomoAccuracy=0.970550576184379;ImpMinorAccuracy=0.85981308411215;DUP\tGT:AD:GL\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0:2,0:0,6,55\t0/0::\t0/0::\t1/1::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/1:1,3:71,0,16\t0/0::\t0/0::\t0/0::\t0/1:1,1:22,0,22\t0/0:3,0:0,9,83\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/1:1,6:145,0,7\t1/1:0,20:555,60,0\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0:7,0:0,21,194\t0/0::\t1/1:0,1:28,3,0\t0/0::\t0/0:3,0:0,9,83\t0/0::\t1/1:0,1:28,3,0\t0/0::\t0/0::\t0/0::\t1/1:0,1:28,3,0\t0/0::\t0/0::\t0/0::\t0/0:3,0:0,9,83\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0:1,0:0,3,28\t0/0:2,0:0,6,55\t0/0::\t0/0:3,0:0,9,83\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t./.::\t0/0::\t./.::\t0/0::\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0:8,0:0,24,222\t0/1:1,3:71,0,16\t0/0:1,0:0,3,28\t0/0::\t0/1:1,4:96,0,13\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t1/1::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:2,0:0,6,55\t0/0:2,0:0,6,55\t0/0::\t0/0:2,0:0,6,55\t0/0:5,0:0,15,139\t0/0:1,0:0,3,28\t0/0:3,0:0,9,83\t0/0:1,0:0,3,28\t0/0:4,0:0,12,111\t0/0::\t0/0::\t0/0::\t1/1:0,1:28,3,0\t1/1::\t1/1:0,2:55,6,0\t0/0:1,0:0,3,28\t0/0:2,0:0,6,55\t0/0:1,0:0,3,28\t0/0::\t0/0::\t./.::\t0/0::\t0/0:1,0:0,3,28\t0/0:1,0:0,3,28\t0/0:2,0:0,6,55\t0/0:1,0:0,3,28\t0/0::\t0/0:8,0:0,24,222\t0/0:2,0:0,6,55\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t0/0:6,0:0,18,166\t0/0::\t0/0:1,0:0,3,28\t0/0:3,0:0,9,83\t1/1:0,7:194,21,0\t0/0:1,0:0,3,28\t1/1:0,1:28,3,0\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0:2,0:0,6,55\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t1/1:0,2:55,6,0\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t1/1::\t0/0:1,0:0,3,28\t0/0::\t0/1:1,2:46,0,19\t0/0:1,0:0,3,28\t0/0:1,0:0,3,28\t0/0:1,0:0,3,28\t1/1:0,2:55,6,0\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t1/1:0,1:28,3,0\t0/0:2,0:0,6,55\t0/0::\t0/0:1,0:0,3,28\t0/0::\t1/1:0,3:83,9,0\t0/0::\t0/0:3,0:0,9,83\t0/0::\t0/0:3,0:0,9,83\t0/0:1,0:0,3,28\t0/0:1,0:0,3,28\t0/0::"
[2] "1\t21000274\t1-20689304\tC\tA\t.\tPASS\tDP=961;NZ=1181;AD=878,83;AC=153;AQ=35,32;GN=1098,13,70;HT=13;EF=1.59;PV=0;MAF=0.065;MAF0=0.068;LLD;INHMP311;ImpHomoAccuracy=0.981186685962373;ImpMinorAccuracy=0.962962962962963;DUP\tGT:AD:GL\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t1/1::\t0/0::\t0/0:2,0:0,6,55\t0/0::\t1/1::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/1:6,3:56,0,139\t1/1:0,13:361,39,0\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0::\t1/1:0,1:28,3,0\t0/0::\t0/0:3,0:0,9,83\t0/0::\t./.::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t1/1::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0:2,0:0,6,55\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0:3,0:0,9,83\t0/0::\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t./.::\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:3,0:0,9,83\t0/0:15,0:0,45,416\t1/1:0,1:28,3,0\t0/0:1,0:0,3,28\t0/0::\t0/1:12,8:162,0,273\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t1/1::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0:2,0:0,6,55\t0/0:4,0:0,12,111\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t1/1:0,1:28,3,0\t1/1::\t1/1::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t./.::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0:4,0:0,12,111\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:15,0:0,45,416\t0/0::\t0/0::\t0/0:3,0:0,9,83\t1/1:0,5:139,15,0\t0/0:3,0:0,9,83\t1/1::\t0/0:2,0:0,6,55\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:4,0:0,12,111\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0:3,0:0,9,83\t0/0:3,0:0,9,83\t0/0:2,0:0,6,55\t0/0:2,0:0,6,55\t0/0::\t0/1:1,2:46,0,19\t0/0:3,0:0,9,83\t0/0::\t0/0:2,0:0,6,55\t0/0:2,0:0,6,55\t0/0:1,0:0,3,28\t0/0:1,0:0,3,28\t0/0:1,0:0,3,28\t0/1:1,1:22,0,22\t0/0:2,0:0,6,55\t0/0:2,0:0,6,55\t./.::\t1/1:0,1:28,3,0\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t1/1:0,2:55,6,0\t0/0:2,0:0,6,55\t0/0:2,0:0,6,55\t0/0:3,0:0,9,83\t0/0::\t0/1:1,3:71,0,16\t0/0:1,0:0,3,28\t0/0:5,0:0,15,139\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::"
head(testlines[!testLLD], n = 2) # lines that should not have LLD
[1] "1\t21000162\t1-20689192\tG\tT\t.\tPASS\tDP=853;NZ=1203;AD=844,9;AC=14;AQ=34,34;GN=1195,2,6;HT=2;EF=1;PV=0.001;MAF=0.006;MAF0=0.02;IBD1;ImpHomoAccuracy=0.983941605839416;ImpMinorAccuracy=0\tGT:AD:GL\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0:6,0:0,18,166\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0:3,0:0,9,83\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:5,0:0,15,139\t0/0:21,0:0,63,583\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0:5,0:0,15,139\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t0/0:3,0:0,9,83\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0::\t./.::\t0/0:1,0:0,3,28\t0/0::\t0/0:4,0:0,12,111\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:5,0:0,15,139\t0/0:4,0:0,12,111\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:6,0:0,18,166\t0/0:2,0:0,6,55\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0:3,0:0,9,83\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0:1,0:0,3,28\t0/0:1,0:0,3,28\t0/0:1,0:0,3,28\t0/0::\t0/0:3,0:0,9,83\t0/0::\t0/0::\t0/0::\t0/0:4,0:0,12,111\t0/0:1,0:0,3,28\t0/0:2,0:0,6,55\t0/0:1,0:0,3,28\t0/0:1,0:0,3,28\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0:4,0:0,12,111\t0/0:2,0:0,6,55\t0/0:2,0:0,6,55\t0/0::\t0/0:9,0:0,27,250\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0:6,0:0,18,166\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0:5,0:0,15,139\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/1:2,1:19,0,46\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0:3,0:0,9,83\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0:3,0:0,9,83\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::"
[2] "1\t21000190\t1-20689220\tG\tA\t.\tPASS\tDP=857;NZ=1203;AD=847,10;AC=15;AQ=33,32;GN=1194,3,6;HT=3;EF=1.23;PV=0.002;MAF=0.006;MAF0=0.019;ImpHomoAccuracy=0.988387096774194;ImpMinorAccuracy=0.307692307692308\tGT:AD:GL\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0:4,0:0,12,111\t0/0::\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0:3,0:0,9,83\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0:7,0:0,21,194\t0/0:20,0:0,60,555\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0:7,0:0,21,194\t0/0::\t0/0:3,0:0,9,83\t0/0::\t0/0:3,0:0,9,83\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:3,0:0,9,83\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0:3,0:0,9,83\t0/0:4,0:0,12,111\t0/0::\t0/0:3,0:0,9,83\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0:5,0:0,15,139\t0/0:4,0:0,12,111\t0/0::\t0/0::\t0/0:5,0:0,15,139\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:2,0:0,6,55\t0/0:2,0:0,6,55\t0/0:1,0:0,3,28\t0/1:1,2:46,0,19\t0/0:2,0:0,6,55\t0/0:1,0:0,3,28\t0/0:1,0:0,3,28\t0/0:1,0:0,3,28\t0/0:6,0:0,18,166\t0/0::\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0:2,0:0,6,55\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0:2,0:0,6,55\t0/0:2,0:0,6,55\t0/0:1,0:0,3,28\t0/0::\t0/0:5,0:0,15,139\t0/0:2,0:0,6,55\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t0/0:7,0:0,21,194\t0/0::\t0/0:1,0:0,3,28\t0/0:1,0:0,3,28\t0/0:3,0:0,9,83\t0/0:1,0:0,3,28\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t0/0::\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:3,0:0,9,83\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0:3,0:0,9,83\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0::\t0/0:1,0:0,3,28\t0/0:2,0:0,6,55\t0/0::\t0/0::\t0/0::\t0/0:2,0:0,6,55\t0/0::\t0/0:3,0:0,9,83\t0/0::\t0/0:3,0:0,9,83\t0/0:1,0:0,3,28\t0/0:1,0:0,3,28\t0/0::"
grepl
If you are confused about what
grepldoes, try:grepl("a", c("cat", "dog", "banana", "ACGT"))
When the LLD flag is present, we see it after MAF0. Now let’s build our
command into a function, and make a similar one for DUP.
LLD_yes <- function(lines){
grepl("LLD", lines)
}
DUP_no <- function(lines){
!grepl("DUP", lines)
}
head(LLD_yes(testlines))
[1] FALSE FALSE TRUE TRUE FALSE TRUE
head(info(test)$LLD)
[1] FALSE FALSE TRUE TRUE FALSE TRUE
head(DUP_no(testlines))
[1] TRUE TRUE FALSE FALSE TRUE FALSE
head(info(test)$DUP)
[1] FALSE FALSE TRUE TRUE FALSE TRUE
These tests on the first six variants show that the functions are behaving as expected.
Now that we have build these three functions, we can use them for filtering the
VCF. Functions that act on plain text go into the prefilters argument, and
functions that act on imported data go into the filters argument. We’ll set
index to TRUE so that the output will already be compressed and indexed for
our use.
filterVcf(myvcf, genome = "Zm-B73-4.0",
destination = "data/hmp321_agpv4_chr1_subset_filtered.vcf",
prefilters = FilterRules(list(LLD_yes, DUP_no)),
filters = FilterRules(list(maf01)),
index = TRUE)
starting prefilter
prefiltering 120304 records
prefiltered to C:\Users\lvclark\AppData\Local\Temp\RtmpYJMA8u\file28341ebc6331
prefilter compressing and indexing C:\Users\lvclark\AppData\Local\Temp\RtmpYJMA8u\file28341ebc6331
starting filter
filtering 41500 records
completed filtering
compressing and indexing data/hmp321_agpv4_chr1_subset_filtered.vcf
If you look at the data directory, you should see the new file and its
index now.
Challenge
Make a filtering function that would only retain markers where the total read depth was over 2000.
Solution
DP2000 <- function(vcf){ info(vcf)$DP > 2000 }
Bonus challenge
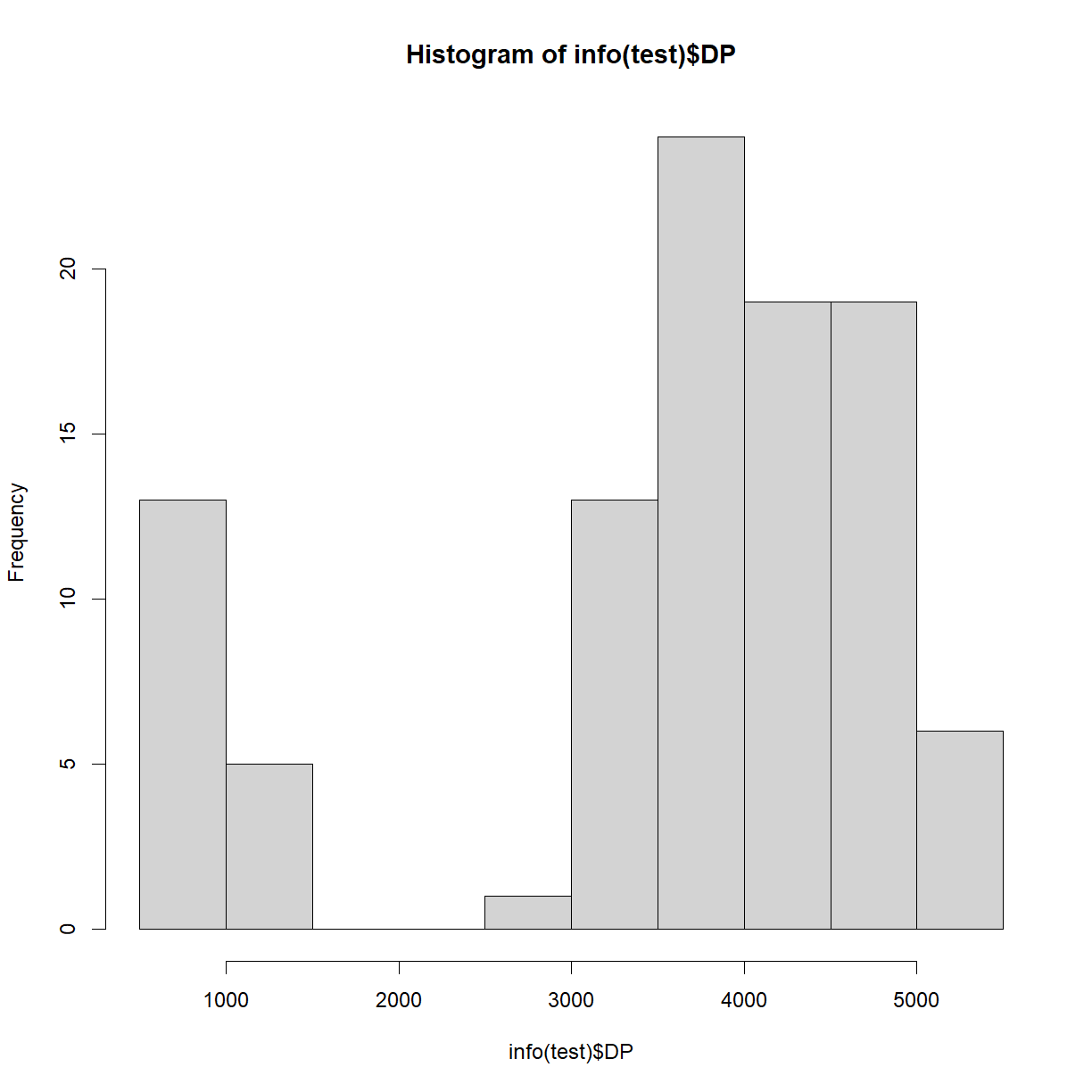
Use your knowledge of R to plot the distribution of total read depth in the first 100 markers (the
testobject).Solution
One quick solution is:
hist(info(test)$DP)
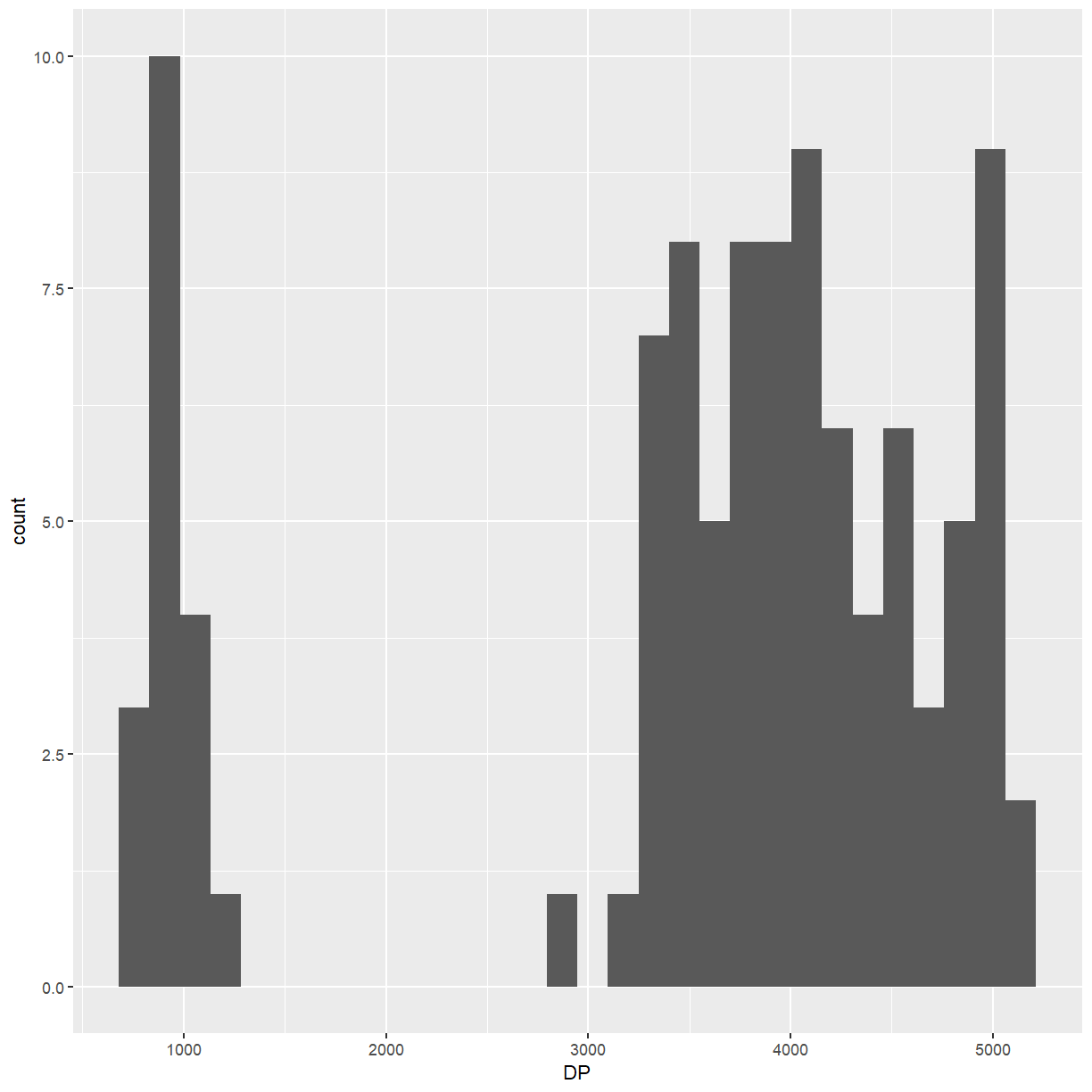
But there are many other possible ways to do it, such as using
ggplot2. Note thatggplotunderstandsdata.frames andtibbles but notDataFrames, so you’ll have to convertinfo(test).library(ggplot2) info(test) %>% as.data.frame() %>% ggplot(mapping = aes(x = DP)) + geom_histogram()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
Custom VCF import
Ok, now we have previewed and filtered the VCF, and we would like to actually
read in data and begin our analysis. But we still don’t want to read the whole
thing into memory. We probably want the genotypes in the GT field, but don’t
care about the AD or GL fields. In the INFO table, we definitely don’t
need LLD or DUP any more since we filtered on them, and in fact maybe the
only fields that we care about are DP and MAF. It’s also possible that we
only want to import certain samples or certain regions.
Let’s say that we know we want to omit samples “32” and “98F1”. We’ll make a vector of sample names to keep.
keep_sam <- all_sam[!all_sam %in% c("32", "98F1")]
We can also just import particular regions. Since this is a small example
dataset, I can tell you that it only covers chromosome 1 from 21 to 24 Mb.
To only import particular regions within that, we’ll make a GRanges.
keep_regions <- GRanges(seqnames = "1",
ranges = IRanges(start = c(21.4e6, 22.9e6),
end = c(22.3e6, 23.8e6)))
names(keep_regions) <- c("Region_1", "Region_2")
keep_regions
GRanges object with 2 ranges and 0 metadata columns:
seqnames ranges strand
<Rle> <IRanges> <Rle>
Region_1 1 21400000-22300000 *
Region_2 1 22900000-23800000 *
-------
seqinfo: 1 sequence from an unspecified genome; no seqlengths
We can do all of our selection using ScanVcfParam. Take a look at the help
page for this function. If we want to keep all fields in a certain category, we
can leave the respective argument at the default.
Below we’ll use:
infoto indicate whichINFOcolumns to keepgenoto indicate whichFORMATcolumns to keepsamplesto indicate which samples to keepwhichto indicate genomic regions to keep.
svp <- ScanVcfParam(info = c("DP", "MAF"), geno = "GT",
samples = keep_sam, which = keep_regions)
Challenge: Ignore QUAL and FILTER
The
QUALandFILTERcolumns don’t contain any useful information in this file, so we don’t need them. Modify oursvpobject to omit them.Solution
We see from the help file that
fixedcan containALT,QUAL, andFILTER. So, we can just set it toALT. We can modify our existing object like so:vcfFixed(svp) <- "ALT"Or we can just remake the object:
svp <- ScanVcfParam(info = c("DP", "MAF"), geno = "GT", samples = keep_sam, which = keep_regions, fixed = "ALT")
Now, let’s import our VCF, just keeping the data that we want.
We’ll send all of our parameters to the param argument.
Since we are specifying genomic regions, we also need to make sure
those chromosome names are present in the genome argument.
myvcf3 <- VcfFile("data/hmp321_agpv4_chr1_subset_filtered.vcf.bgz")
mydata <- readVcf(myvcf3, param = svp, genome = seqlevels(keep_regions))
mydata
class: CollapsedVCF
dim: 24528 198
rowRanges(vcf):
GRanges with 3 metadata columns: paramRangeID, REF, ALT
info(vcf):
DataFrame with 2 columns: DP, MAF
info(header(vcf)):
Number Type Description
DP 1 Integer Total Depth
MAF 1 Float Minor allele frequency
geno(vcf):
List of length 1: GT
geno(header(vcf)):
Number Type Description
GT 1 String Genotype
One handy thing is that rowRanges now has a column called paramRangeID
to indicate which genomic range that SNP corresponds to.
rowRanges(mydata)
GRanges object with 24528 ranges and 3 metadata columns:
seqnames ranges strand | paramRangeID REF
<Rle> <IRanges> <Rle> | <factor> <DNAStringSet>
1-21094080 1 21400101 * | Region_1 A
1-21094095 1 21400116 * | Region_1 C
1-21094140 1 21400161 * | Region_1 A
1-21094172 1 21400193 * | Region_1 G
1-21094216 1 21400237 * | Region_1 C
... ... ... ... . ... ...
1-23395014 1 23784652 * | Region_2 C
1-23395152 1 23784790 * | Region_2 C
1-23395224 1 23784862 * | Region_2 A
1-23395794 1 23785432 * | Region_2 G
1-23395848 1 23785486 * | Region_2 C
ALT
<CharacterList>
1-21094080 G
1-21094095 G
1-21094140 G
1-21094172 T
1-21094216 T
... ...
1-23395014 T
1-23395152 T
1-23395224 G
1-23395794 A
1-23395848 T
-------
seqinfo: 1 sequence from 1 genome; no seqlengths
In the next episode we’ll start some analysis of the SNP genotypes.
Key Points
Index the VCF file with indexTabix if you plan to only import certain ranges.
Use filterVcf to filter variants to a new file without importing data into R.
Use ScanVcfParam to specify which fields, samples, and genomic ranges you want to import.