Nextflow configuration
Last updated on 2024-12-04 | Edit this page
Estimated time: 45 minutes
Overview
Questions
- What is the difference between the workflow implementation and the workflow configuration?
- How do I configure a Nextflow workflow?
- How do I assign different resources to different processes?
- How do I separate and provide configuration for different computational systems?
- How do I change configuration settings from the default settings provided by the workflow?
Objectives
- Understand the difference between workflow implementation and configuration.
- Understand the difference between configuring Nextflow and a Nextflow script.
- Create a Nextflow configuration file.
- Understand what a configuration scope is.
- Be able to assign resources to a process.
- Be able to refine configuration settings using process selectors.
- Be able to group configurations into profiles for use with different computer infrastructures.
- Be able to override existing settings.
- Be able to inspect configuration settings before running a workflow.
Nextflow configuration
A key Nextflow feature is the ability to decouple the workflow implementation, which describes the flow of data and operations to perform on that data, from the configuration settings required by the underlying execution platform. This enables the workflow to be portable, allowing it to run on different computational platforms such as an institutional HPC or cloud infrastructure, without needing to modify the workflow implementation.
We have seen earlier that it is possible to provide a
process with directives. These directives are process
specific configuration settings. Similarly, we have also provided
parameters to our workflow which are parameter configuration settings.
These configuration settings can be separated from the workflow
implementation, into a configuration file.
Configuration files
Settings in a configuration file are sets of name-value pairs
(name = value). The name is a specific
property to set, while the value can be anything you can
assign to a variable (see nextflow
scripting), for example, strings, booleans, or other variables. It
is also possible to access any variable defined in the host environment
such as $PATH, $HOME, $PWD,
etc.
Accessing variables in your configuration file
Settings are also partitioned into scopes, which govern the behaviour
of different elements of the workflow. For example, workflow parameters
are governed from the params scope, while process
directives are governed from the process scope. A full list
of the available scopes can be found in the documentation.
It is also possible to define your own scope.
Configuration settings for a workflow are often stored in the file
nextflow.config which is in the same directory as the
workflow script. Configuration can be written in either of two ways. The
first is using dot notation, and the second is using brace notation.
Both forms of notation can be used in the same configuration file.
An example of dot notation:
GROOVY
params.input = '' // The workflow parameter "input" is assigned an empty string to use as a default value
params.outdir = './results' // The workflow parameter "outdir" is assigned the value './results' to use by default.An example of brace notation:
Configuration files can also be separated into multiple files and
included into another using the includeConfig
statement.
How configuration files are combined
Configuration settings can be spread across several files. This also allows settings to be overridden by other configuration files. The priority of a setting is determined by the following order, ranked from highest to lowest.
- Parameters specified on the command line
(
--param_name value). - Parameters provided using the
-params-fileoption. - Config file specified using the
-cmy_config option. - The config file named
nextflow.configin the current directory. - The config file named
nextflow.configin the workflow project directory ($projectDir: the directory where the script to be run is located). - The config file
$HOME/.nextflow/config. - Values defined within the workflow script itself (e.g.,
main.nf).
If configuration is provided by more than one of these methods, configuration is merged giving higher priority to configuration provided higher in the list.
Existing configuration can be completely ignored by using
-C <custom.config> to use only configuration provided
in the custom.config file.
Configuring Nextflow vs Configuring a Nextflow workflow
Parameters starting with a single dash - (e.g.,
-c my_config.config) are configuration options for
nextflow, while parameters starting with a double dash
-- (e.g., --outdir) are workflow parameters
defined in the params scope.
The majority of Nextflow configuration settings must be provided on
the command-line, however a handful of settings can also be provided
within a configuration file, such as
workdir = '/path/to/work/dir'
(-w /path/to/work/dir) or resume = true
(-resume), and do not belong to a configuration scope.
Determine script output
Determine the outcome of the following script executions. Given the
script print_message.nf:
GROOVY
nextflow.enable.dsl = 2
params.message = 'hello'
workflow {
PRINT_MESSAGE(params.message)
}
process PRINT_MESSAGE {
echo true
input:
val my_message
script:
"""
echo $my_message
"""
}and configuration (print_message.config):
params.message = 'Are you tired?'What is the outcome of the following commands?
nextflow run print_message.nfnextflow run print_message.nf --message '¿Que tal?'nextflow run print_message.nf -c print_message.confignextflow run print_message.nf -c print_message.config --message '¿Que tal?'
- ‘hello’ - Workflow script uses the value in
print_message.nf - ‘¿Que tal?’ - The command-line parameter overrides the script setting.
- ‘Are you tired?’ - The configuration overrides the script setting
- ‘¿Que tal?’ - The command-line parameter overrides both the script and configuration settings.
Configuring process behaviour
Earlier we saw that process directives allow the
specification of settings for the task execution such as
cpus, memory, conda and other
resources in the pipeline script. This is useful when prototyping a
small workflow script, however this ties the configuration to the
workflow, making it less portable. A good practice is to separate the
process configuration settings into another file.
The process configuration scope allows the setting of
any process directives in the Nextflow configuration file.
For example:
GROOVY
// nextflow.config
process {
cpus = 2
memory = 8.GB
time = '1 hour'
publishDir = [ path: params.outdir, mode: 'copy' ]
}Unit values
Memory and time duration units can be specified either using a string based notation in which the digit(s) and the unit can be separated by a space character, or by using the numeric notation in which the digit(s) and the unit are separated by a dot character and not enclosed by quote characters.
| String syntax | Numeric syntax | Value |
|---|---|---|
| ‘10 KB’ | 10.KB | 10240 bytes |
| ‘500 MB’ | 500.MB | 524288000 bytes |
| ‘1 min’ | 1.min | 60 seconds |
| ‘1 hour 25 sec’ | - | 1 hour and 25 seconds |
These settings are applied to all processes in the workflow. A process selector can be used to apply the configuration to a specific process or group of processes.
Process selectors
The resources for a specific process can be defined using
withName: followed by the process name e.g.,
'FASTQC', and the directives within curly braces. For
example, we can specify different cpus and
memory resources for the processes INDEX and
FASTQC as follows:
GROOVY
// process_resources.config
process {
withName: INDEX {
cpus = 4
memory = 8.GB
}
withName: FASTQC {
cpus = 2
memory = 4.GB
}
}When a workflow has many processes, it is inconvenient to specify
directives for all processes individually, especially if directives are
repeated for groups of processes. A helpful strategy is to annotate the
processes using the label directive (processes can have
multiple labels). The withLabel selector then allows the
configuration of all processes annotated with a specific label, as shown
below:
GROOVY
// configuration_process_labels.nf
process P1 {
label "big_mem"
script:
"""
echo P1: Using $task.cpus cpus and $task.memory memory.
"""
}
process P2 {
label "big_mem"
script:
"""
echo P2: Using $task.cpus cpus and $task.memory memory.
"""
}
workflow {
P1()
P2()
}GROOVY
// configuration_process-labels.config
process {
withLabel: big_mem {
cpus = 16
memory = 64.GB
}
}Another strategy is to use process selector expressions. Both
withName: and withLabel: allow the use of
regular expressions to apply the same configuration to all processes
matching a pattern. Regular expressions must be quoted, unlike simple
process names or labels.
- The
|matches either-or, e.g.,withName: 'INDEX|FASTQC'applies the configuration to any process matching the nameINDEXorFASTQC. - The
!inverts a selector, e.g.,withLabel: '!small_mem'applies the configuration to any process without thesmall_memlabel. - The
.*matches any number of characters, e.g.,withName: 'NFCORE_RNASEQ:RNA_SEQ:BAM_SORT:.*'matches all processes of the workflowNFCORE_RNASEQ:RNA_SEQ:BAM_SORT.
A regular expression cheat-sheet can be found here if you would like to write more expressive expressions.
Selector priority
When mixing generic process configuration and selectors, the following priority rules are applied (from highest to lowest):
-
withNameselector definition. -
withLabelselector definition. - Process specific directive defined in the workflow script.
- Process generic
processconfiguration.
Process selectors
Create a Nextflow config, process-selector.config,
specifying different cpus and memory resources
for the two processes P1 (cpus 1 and memory 2.GB) and
P2 (cpus 2 and memory 1.GB), where P1 and
P2 are defined as follows:
GROOVY
// process-selector.config
process {
withName: P1 {
cpus = 1
memory = 2.GB
}
withName: P2 {
cpus = 2
memory = 1.GB
}
}OUTPUT
N E X T F L O W ~ version 21.04.0
Launching `process-selector.nf` [clever_borg] -
revision: e765b9e62d
executor > local (2)
[de/86cef0] process > P1 [100%] 1 of 1 ✔
[bf/8b332e] process > P2 [100%] 1 of 1 ✔
P2: Using 2 cpus and 1 GB memory.
P1: Using 1 cpus and 2 GB memory.Dynamic expressions
A common scenario is that configuration settings may depend on the
data being processed. Such settings can be dynamically expressed using a
closure. For example, we can specify the memory required as
a multiple of the number of cpus. Similarly, we can publish
results to a subfolder based on the sample name.
Configuring execution platforms
Nextflow supports a wide range of execution platforms, from running locally, to running on HPC clusters or cloud infrastructures. See https://www.nextflow.io/docs/latest/executor.html for the full list of supported executors.
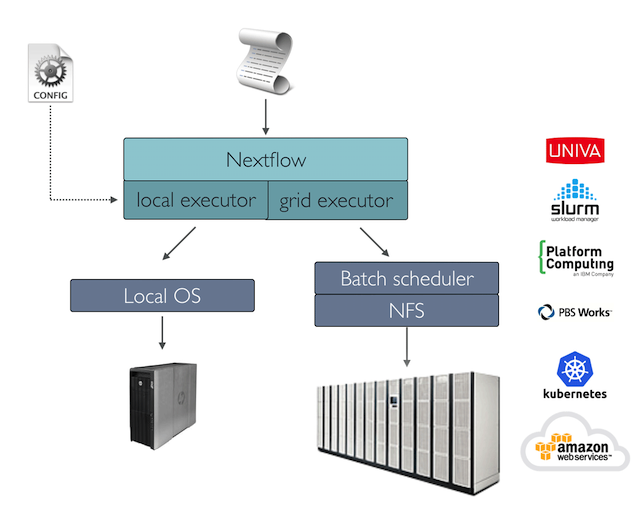
The default executor configuration is defined within the
executor scope (https://www.nextflow.io/docs/latest/config.html#scope-executor).
For example, in the config below we specify the executor as Sun Grid
Engine, sge and the number of tasks the executor will
handle in a parallel manner (queueSize) to 10.
The process.executor directive allows you to override
the executor to be used by a specific process. This can be useful, for
example, when there are short running tasks that can be run locally, and
are unsuitable for submission to HPC executors (check for guidelines on
best practice use of your execution system). Other process directives
such as process.clusterOptions, process.queue,
and process.machineType can be also be used to further
configure processes depending on the executor used.
Configuring software requirements
An important feature of Nextflow is the ability to manage software
using different technologies. It supports the Conda package management
system, and container engines such as Docker, Singularity, Podman,
Charliecloud, and Shifter. These technologies allow one to package tools
and their dependencies into a software environment such that the tools
will always work as long as the environment can be loaded. This
facilitates portable and reproducible workflows. Software environment
specification is managed from the process scope, allowing
the use of process selectors to manage which processes load which
software environment. Each technology also has its own scope to provide
further technology specific configuration settings.
Software configuration using Conda
Conda is a software package and environment management system that runs on Linux, Windows, and Mac OS. Software packages are bundled into Conda environments along with their dependencies for a particular operating system (Not all software is supported on all operating systems). Software packages are tied to conda channels, for example, bioinformatic software packages are found and installed from the BioConda channel.
A Conda environment can be configured in several ways:
- Provide a path to an existing Conda environment.
- Provide a path to a Conda environment specification file (written in YAML).
- Specify the software package(s) using the
<channel>::<package_name>=<version>syntax (separated by spaces), which then builds the Conda environment when the process is run.
GROOVY
process {
conda = "/home/user/miniconda3/envs/my_conda_env"
withName: FASTQC {
conda = "environment.yml"
}
withName: SALMON {
conda = "bioconda::salmon=1.5.2"
}
}There is an optional conda scope which allows you to
control the creation of a Conda environment by the Conda package
manager. For example, conda.cacheDir specifies the path
where the Conda environments are stored. By default this is in
conda folder of the work directory.
This following exercise necessitates the installation of conda, a package and environment management software, on your system for downloading and installing the FASTP software. If conda is not installed, then this exercise should be omitted.
Define a software requirement in the configuration file using conda
We are going to define a software requirement using the “conda” directive.
The software we are going to uses is fastp, a tool used
for fast processing of next-generation sequencing data (like RNA or DNA
sequences).
Each time the process is called we are going to run
fastp -A -i ${read} -o out.fq 2>&1
-
fastpis the main command that invokes the fastp program. - -A: option tells fastp to automatically detect and trim adapters. Adapters are short, artificially added sequences used in sequencing that are not part of the target DNA or RNA. -i ${read}:
- The
-iflag specifies the input file.${read}represents a nextflow variable that contains the path to the sequencing reads. -
-o: The-oflag specifies the output file. In this case, the processed sequencing data will be written to a file namedout.fq. -
2>&1: This is a shell redirection command, it means that both the regular output and error messages will be sent to the console.
Create a config file for the Nextflow script
configuration_fastp.nf.Add a conda directive for the process name
FASTPthat includes the bioconda packagefastp, version 0.12.4-0.
Hint You can specify the conda packages using the
syntax
<channel>::<package_name>=<version>
e.g. bioconda::salmon=1.5.2
- Run the Nextflow script
configure_fastp.nfwith the configuration file using the-coption.
OUTPUT
N E X T F L O W ~ version 21.04.0
Launching `configuration_fastp.nf` [berserk_jepsen] - revision: 28fadd2486
executor > local (1)
[c1/c207d5] process > FASTP (1) [100%] 1 of 1 ✔
Creating Conda env: bioconda::fastp=0.12.4-0 [cache /home/training/work/conda/env-a7a3a0d820eb46bc41ebf4f72d955e5f]
ref1_1.fq.gz 58708
Read1 before filtering:
total reads: 14677
total bases: 1482377
Q20 bases: 1466210(98.9094%)
Q30 bases: 1415997(95.5221%)
Read1 after filtering:
total reads: 14671
total bases: 1481771
Q20 bases: 1465900(98.9289%)
Q30 bases: 1415769(95.5457%)
Filtering result:
reads passed filter: 14671
reads failed due to low quality: 6
reads failed due to too many N: 0
reads failed due to too short: 0
JSON report: fastp.json
HTML report: fastp.htmlSoftware configuration using Docker
Docker is a container technology. Container images are lightweight, standalone, executable package of software that includes everything needed to run an application: code, runtime, system tools, system libraries and settings. Containerized software is intended to run the same regardless of the underlying infrastructure, unlike other package management technologies which are operating system dependant (See the published article on Nextflow). For each container image used, Nextflow uses Docker to spawn an independent and isolated container instance for each process task.
To use Docker, we must provide a container image path using the
process.container directive, and also enable docker in the
docker scope, docker.enabled = true. A container image path
takes the form
(protocol://)registry/repository/image:version--build. By
default, Docker containers run software using a privileged user. This
can cause issues, and so it is also a good idea to supply your user and
group via the docker.runOptions.
The Docker run option -u $(id -u):$(id -g) is used to
specify the user and group IDs (UID and GID) that the container should
use when running. This option ensures that the processes inside the
container run with the same UID and GID as the user executing the Docker
command on the host system.
Software configuration using Singularity
Singularity is another container technology, commonly used on HPC
clusters. It is different to Docker in several ways. The primary
differences are that processes are run as the user, and certain
directories are automatically “mounted” (made available) in the
container instance. Singularity also supports building Singularity
images from Docker images, allowing Docker image paths to be used as
values for process.container.
Singularity is enabled in a similar manner to Docker. A container
image path must be provided using process.container and
singularity enabled using singularity.enabled = true.
GROOVY
process.container = 'https://depot.galaxyproject.org/singularity/salmon:1.5.2--h84f40af_0'
singularity.enabled = trueContainer protocols
The following protocols are supported:
-
docker://: download the container image from the Docker Hub and convert it to the Singularity format (default). -
library://: download the container image from the Singularity Library service. -
shub://: download the container image from the Singularity Hub. -
docker-daemon://: pull the container image from a local Docker installation and convert it to a Singularity image file. -
https://: download the singularity image from the given URL. -
file://: use a singularity image on local computer storage.
Configuration profiles
One of the most powerful features of Nextflow configuration is to
predefine multiple configurations or profiles for different
execution platforms. This allows a group of predefined settings to be
called with a short invocation,
-profile <profile name>.
Configuration profiles are defined in the profiles
scope, which group the attributes that belong to the same profile using
a common prefix.
GROOVY
//configuration_profiles.config
profiles {
standard {
params.genome = '/local/path/ref.fasta'
process.executor = 'local'
}
cluster {
params.genome = '/data/stared/ref.fasta'
process.executor = 'sge'
process.queue = 'long'
process.memory = '10GB'
process.conda = '/some/path/env.yml'
}
cloud {
params.genome = '/data/stared/ref.fasta'
process.executor = 'awsbatch'
process.container = 'cbcrg/imagex'
docker.enabled = true
}
}This configuration defines three different profiles:
standard, cluster and cloud that
set different process configuration strategies depending on the target
execution platform. By convention the standard profile is implicitly
used when no other profile is specified by the user. To enable a
specific profile use -profile option followed by the
profile name:
Configuration order
Settings from profiles will override general settings in the
configuration file. However, it is also important to remember that
configuration is evaluated in the order it is read in. For example, in
the following example, the publishDir directive will always
take the value ‘results’ even when the profile hpc is used.
This is because the setting is evaluated before Nextflow knows about the
hpc profile. If the publishDir directive is
moved to after the profiles scope, then
publishDir will use the correct value of
params.results.
Key Points
- Nextflow configuration can be managed using a Nextflow configuration file.
- Nextflow configuration files are plain text files containing a set of properties.
- You can define process specific settings, such as cpus and memory,
within the
processscope. - You can assign different resources to different processes using the
process selectors
withNameorwithLabel. - You can define a profile for different configurations using the
profilesscope. These profiles can be selected when launching a pipeline execution by using the-profilecommand-line option - Nextflow configuration settings are evaluated in the order they are read-in.
